|
K562 cell line. 
|
|
UMAP decomposition. 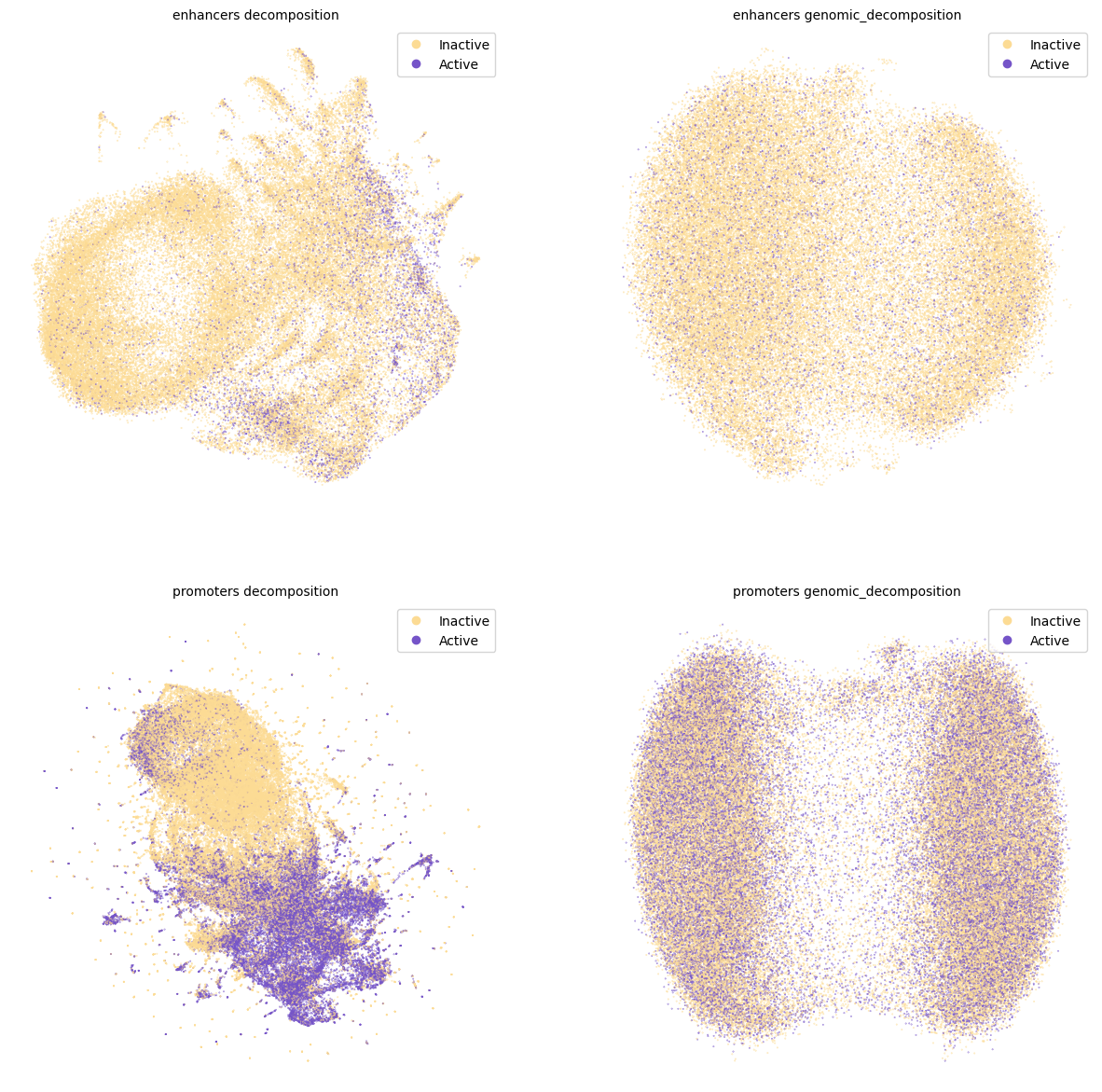
|
|
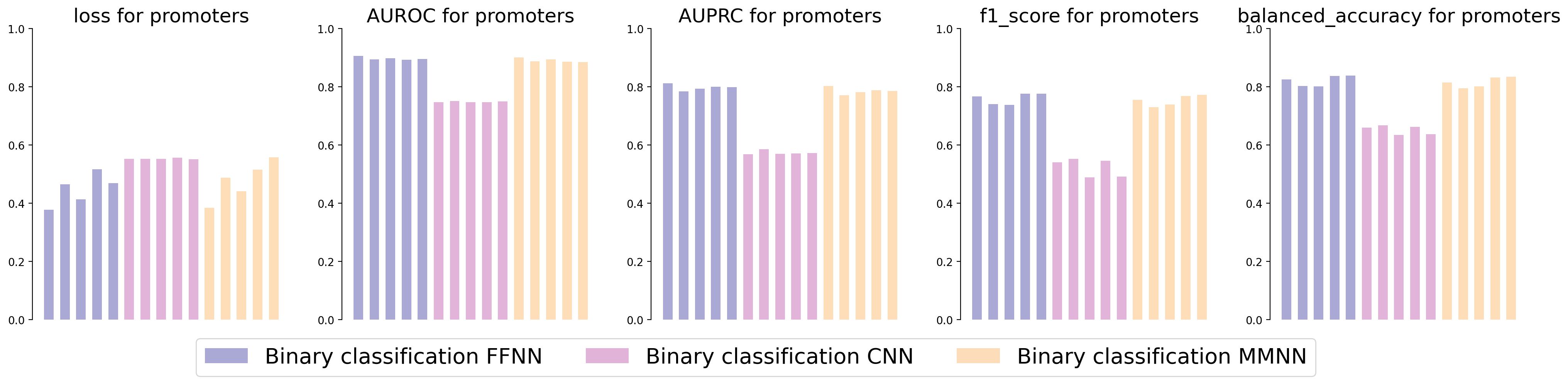
Evaluation metrics. 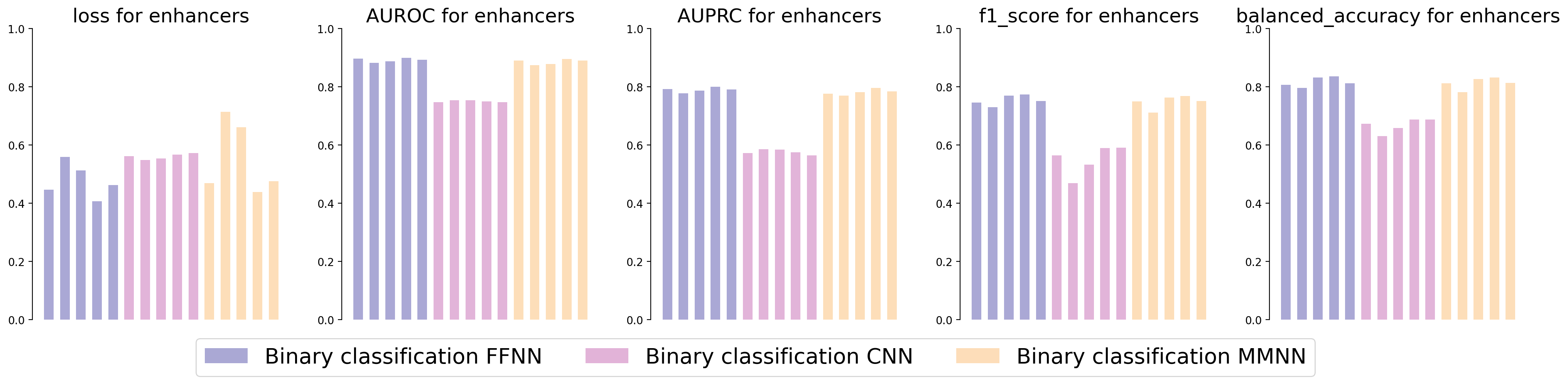
|
Abstract:
To solve the problem of classification of active regulatory regions, we try to predict whether the enhancers and promoters of the cell line K562 are active or inactive. This binary classification problem has been approached in different ways: we explore the possibility of doing it by the implementation of deep learning techniques. First, a FFNN will be trained on the epigenomic data of K652, with levels of activation from FANTOM5. Secondly, a CNN is trained on the reference genomic sequence \emph{hg38}. To fully grasp the complexity of the task, we concatenate the two networks into a MMNN, which uses both type of data to solve the task. Results are close to those of S.O.A., even if the training has been done on limited resources, and only on one cell line. This implies that more tests are needed to determine a statistical relevance.
|
K562 cell line. 
|
|
UMAP decomposition. 
|
|
Evaluation metrics. 

|